The Gene Thicket
TUM Master's Thesis
PDF and CODE
The gene thicket is my Master’s thesis project. The gene thicket is a deep learning-based model, which goal is to build gene regulatory networks that are capable to predict future gene expressions.
What is so amazing about gene regulatory networks?
Gene Regulatory Networks are graphs that describe the regulatory process of a system. Their nodes are genes and their edges represent the regulatory relationship between two genes. The edges have a direction (which gene regulates which other gene), a sign (activation or repression) and a weight (strength of the relationship). If we understand how a system works, then we can develop better products to help people, animals and every other living organism.

The inference of gene regulatory networks (GRNs) has been an area of research for more than twenty years! GRNs are an amazing challenge in Computational Biology since they relate to many different things, such as perturbations, trajectory inference, RNA velocity and much more! We can also be creative and incorporate different types of data.
The gene thicket
The gene thicket is a GRN inference method that predicts the future of each cell incorporating scATAC-seq data as prior, temporal data, a CNN architecture that predicts using only the past, and modified filters of attention to include the signs and magnitude of how the transcription factors affect the target genes.
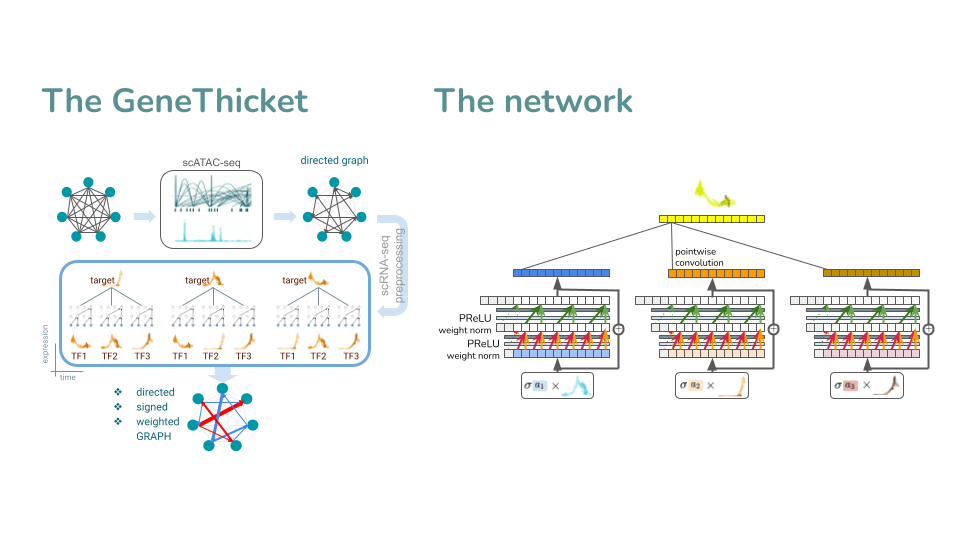
The motivation to build the gene thicket is to have a method that considers non-linear relationships between target genes and transcription factors, such as SCENIC, but that also considers time series data. This is because the change in expression of transcription factors has an impact in the change of expression of target genes, which is observed when the expression profiles are sorted through time.
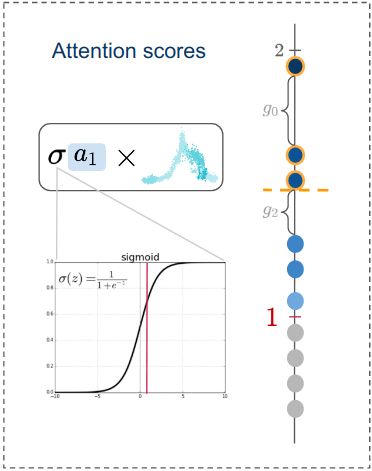
An advantage of having a time series approach is that we can make forecasts about the expression values of target genes using the previous expression values of transcription factors. Then, we could compute displacements and simulate trajectories, in a similar way that CellOracle does. The idea of using deep learning comes from the fact that neural networks are universal non-linear approximators. By using temporal convolutional neural networks with attention, we ensure the scalability and interpretability.
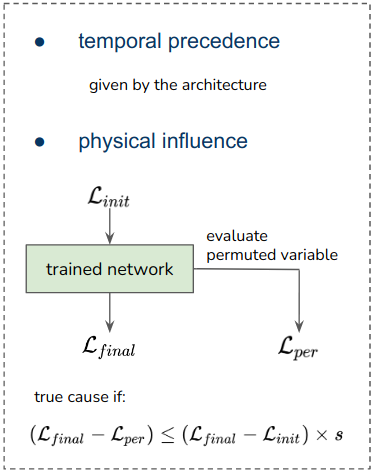
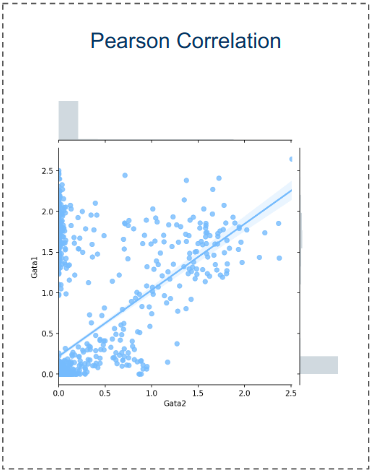
The results
To evaluate the quality of any reconstructed gene regulatory network is extremely challenging, because a ground truth does not exist. Moreover, there are diverse factors that have an impact on experimental data and could affect the interactions in a system. These factors can be primary cell types, environmental conditions, technology platforms and cell lines. To overcome these limitations, I evaluated the model’s performance using synthetic and curated data, and investigated the model’s behavior in real biological data that describes the Pancreatic endocrinogenesis.
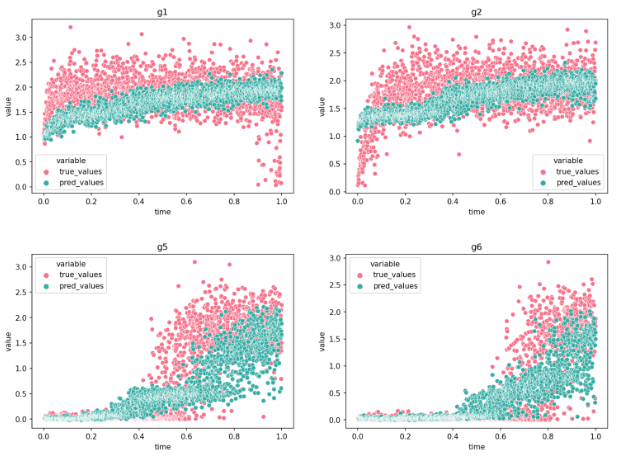
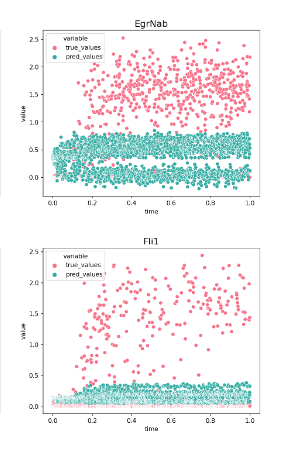
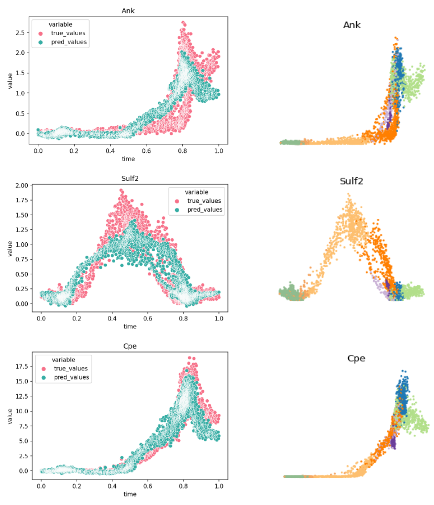
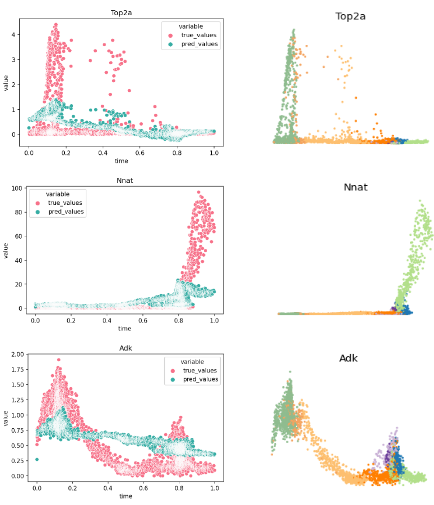
Conclusions and Outlook
- The gene thicket is a first step to iterative GRN-velocity approach.
- We can compute cell displacements using a GRN inference method.
- CNN’s are flexible when computing GRNs.
- able to look at many points in the past
- approximate non-linear trends
- scalable
- can be interpretable (even discover delays)
- We need to consider:
- multiple trajectories
- abrupt changes in gene expression trends
- uncertainty